Research
The area of my research program encompasses the development, function, and homeostasis of T cells as the immune system negotiates commensalism terms with microbiomes of varied composition using Mus musculus as the model system. We focus on two questions
1) How do developing T cells make their functional choice to be CD4+ helpers or CD8+ killers; what regulatory elements are involved and does the microbiome influence that choice?
2) How does the homeostasis of the immune system change in response to a changing microbiome; what genetic features are important, and can prebiotics influence these homeostatic changes?
Research Area 1 – developmental-stage specific regulation of Cd4
During development, precursor cells choose between several potential fates based on the signals they receive from their environment. This decision-making process is a common theme for all multi-cellular organisms. However, this process is especially complex in the development of the immune system, where somatic rearrangement of T cell receptor (TCR) and B cell receptor (BCR) genes results in the outgrowth of a multitude of cells, each with a unique specificity for antigen. For a functional immune system to develop, the mature T lymphocytes must respond to foreign antigens while remaining unresponsive to self-antigens. This discrimination is acquired during T cell development in the thymus through a process called positive selection.
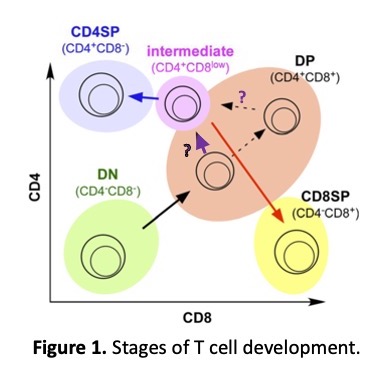
The majority of developing T cells, called thymocytes, which express both CD4 and CD8, must make a lineage decision to become either CD4 or CD8 T cells based on the specificity of their TCR for MHC class I (MHC-I) or class II (MHC-II) loaded with endogenous peptide. Upon receiving a signal via their TCR, CD4+CD8+ double positive (DP) thymocytes downregulate Cd8 and upregulate Cd4 to become CD4+CD8lo intermediate stage (INT) thymocytes (Fig. 1).
As a result, INT cells that use the CD4 coreceptor to stabilize their TCR-MHC-II interaction experience a persistent signal, upregulate Cd4 expression, and commit to the CD4 lineage, while thymocytes that use the CD8 coreceptor to stabilize their TCR-MHC-I interaction experience a cessation of the signal, terminate Cd4 expression, re-express Cd8, and commit to the CD8 lineage (1-3). The regulatory elements involved and the mechanism of Cd4 upregulation in INT cells during positive selection remains unclear.
To understand what regulatory elements in the Cd4 gene ensure CD4 upregulation during positive selection, I continued this line of investigation at Davidson College. Supported by an NIH AREA grant and with the collaboration of numerous undergraduate students in my lab, we were able to identify a novel developmental stage specific element, NCE, in the first intron of the Cd4 gene and demonstrate that it functions as an enhancer both in vitro and in vivo. Using transient transfections of reporter plasmids, we determined that NCE enhances Cd4 promoter function in thymoma cell lines arrested at the INT stage of development, but not the DP stage (Fig. 2A). Consistent with this observation, removing NCE through a CRISPR/Cas9 deletion in INT thymoma cells resulted in lower CD4 expression, while DP thymoma cells were unaffected (Figure 2B). In addition, by stripping pre-existing coreceptors and measuring the rate of CD4 cell surface re-expression over time, we demonstrated that the CD4 rate of expression was reduced in NCE-deleted INT cells, but not NCE-deleted DP cells (Fig. 2C). Taken together, our findings show that NCE has developmental-stage specific enhancer function in vitro at the INT, but not DP, stage of development.
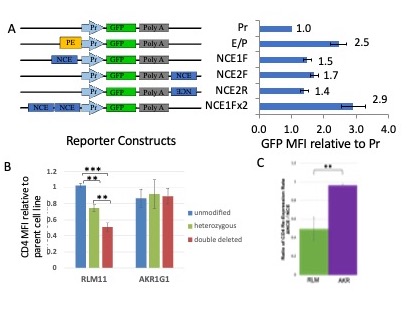
Figure 2. Developmental-stage specific enhancer function of NCE.
A. GFP reporter constructs and NCE enhancer function in transient transfections of the INT-stage arrested RLM11 thymoma. MFI = mean fluorescence intensity, PE = proximal enhancer, Pr = Cd4 promoter. Using a two-tailed comparison to a standard value T-test (Pr MFI set to 1.0) *p<= 0.05 for all measurements.
B. Effect of CRISPR deletion of NCE on CD4 cell surface expression relative to the corresponding parental cell line. AKR1G1 is a DP-stage arrested thymoma cell line. C. After stripping with pronase, CD4 MFI levels were measured by immunostaining and flowcytometry every 2 or 4 hours, linear regression was performed to obtain the rate of re-expression and the ratio of the rate in the absence of NCE to the presence of NCE is shown for RLM and AKR cells.
Another group of research students used Bacterial Artificial Chromosome (BAC) recombineering to generate BAC transgenes that contain the entire Cd4 locus and express EGFP as a reporter instead of CD4 in the presence or absence of NCE. The overall level of EGFP expression in the resulting BAC-transgenic mice was lower in the absence of NCE and instead of increase of EGFP reporter levels from DP to INT thymocytes, there was an actual decrease as determined by RT-qPCR on sorted thymocyte populations, indicating that the developmental-stage specific enhancer function of NCE can be observed in vivo as well.
Research Area 2 – host-microbiome interaction
This project arose from an observation by a student in my immunology lab section that B10.A mice, brought to the Davidson’s conventional animal facility, gradually shifted its CD4:CD8 T cell ratio from one that favors CD4 T cells, considered normal, to one that favors CD8 T cells in a strain-, sex-, and age-dependent manner, unlike their genetically closely related C57BL/10 (B10) mice (Fig. 3).
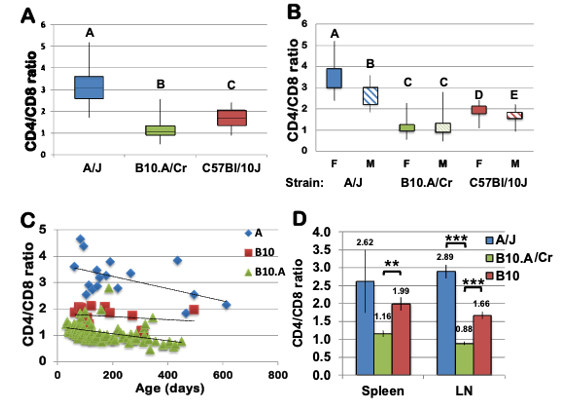
Figure 3. Reduced CD4:CD8 ratio in peripheral blood from B10.A/Cr mice. Percentages of CD4+ and CD8+ cells in tail blood samples were identified by immunostaining with anti-CD4 and anti-CD8 antibodies, followed by flow cytometry.
Three-way ANCOVAmodel was used to analyze differences in the ratio of the frequencies of CD4+ and CD8+ T cells from strains B10.A/Cr, A/J, and C57Bl/10 with age, strain and sex as independent variables (R2=0.73, F=179.4, p<0.0001). Box and whisker plots of A) the CD4:CD8 ratio in each strain and B) sex by strain interaction effect. Different letters indicate statistically significant differences of the means (p<0.0001). F=female, M=male C) Age by strain interaction effect in the model (p=0.0003). D) the CD4:CD8 ratio in Spleen and lymph nodes
The same phenomenon could be observed at other conventional facilities, prompting us to ask two broad sets of questions:
1) Is there a strain-specific genetic issue in T cell generation, maintenance, and function in the periphery, including ability and manner of interacting with ques from the microbiome that could explain the shift in the CD4:CD8 ratio?
2) Are there changes in the environment, especially the microbiome, that drive the shift in the CD4:CD8 ratio and can prebiotics, such as inulin or EGCG, influence the altered T cell homeostasis?
The first question was addressed by an Honor thesis student who determined that the intrinsic properties of T cells do not contribute to this phenotype. T cell development was normal and the mature CD4 and CD8 T cells were able to perform all their usual functions, including respond to TCR stimulation by proliferation, secretion of cytokines, have similar homeostatic proliferation and death rates, and sharing of limiting resources like IL-7. Another set of students investigated more specialized T cell subsets in the periphery, again not finding any substantial differences that could explain the phenotype.
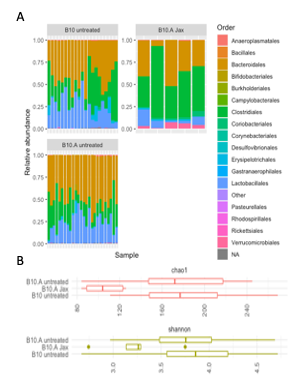
Now we are in the process of looking at the environment of the T cells as determined by the microbiome, the genome, and the transcriptome of the gut. As expected, we see big differences in composition and diversity of the microbiome between an SPF and a conventional facility, however there are no differences in either between B10 and B10.A mice (Fig. 4).
Figure 4. Composition (A) and diversity (B) of the microbiome at Jackson Labs SPF and Davidson’s conventional facility
We find that prebiotic treatment with inulin does not restore the CD4:CD8 ratio, but does arrest its further deterioration, suggesting that microbiome composition is a factor. In the future we plan to investigate the cytokine milieu, the number of B cells, especially IgA-producing ones in the gut, the frequency and identity of IgA-associated bacteria in the colon and correlate that information to a very recently acquired set of colon transcriptomics data on the two mouse strains.
Literature cited:
1. Sarafova, S. D., B. Erman, Q. Yu, F. Van Laethem, T. Guinter, S. O. Sharrow, L. Feigenbaum, K. F. Wildt, W. Ellmeier, and A. Singer. 2005. Modulation of coreceptor transcription during positive selection dictates lineage fate independently of TCR/coreceptor specificity. Immunity 23: 75–87.
2. Adoro, S., B. Erman, S. D. Sarafova, F. Van Laethem, J. H. Park, L. Feigenbaum, and A. Singer. 2008. Targeting CD4 coreceptor expression to postselection thymocytes reveals that CD4/CD8 lineage choice is neither error-prone nor stochastic. J.Immunol. 181: 6975–6983.
3. Sarafova, S. D., F. Van Laethem, S. Adoro, T. Guinter, S. O. Sharrow, L. Feigenbaum, and A. Singer. 2009. Upregulation of CD4 expression during MHC class II-specific positive selection is essential for error-free lineage choice. Immunity 31: 480–490.

Recent Comments